This runs DeepExplainer with the model trained on simualted genomic data from the DeepLIFT repo (https://github.com/kundajelab/deeplift/blob/master/examples/genomics/genomics_simulation.ipynb), using a dynamic reference (i.e. the reference varies depending on the input sequence; in this case, the reference is a collection of dinucleotide-shuffled versions of the input sequence)
[1]:
%matplotlib inline
from __future__ import print_function, division
Pull in the relevant data
[2]:
! [[ ! -f sequences.simdata.gz ]] && wget https://raw.githubusercontent.com/AvantiShri/model_storage/db919b12f750e5844402153233249bb3d24e9e9a/deeplift/genomics/sequences.simdata.gz
! [[ ! -f keras2_conv1d_record_5_model_PQzyq_modelJson.json ]] && wget https://raw.githubusercontent.com/AvantiShri/model_storage/b6e1d69/deeplift/genomics/keras2_conv1d_record_5_model_PQzyq_modelJson.json
! [[ ! -f keras2_conv1d_record_5_model_PQzyq_modelWeights.h5 ]] && wget https://raw.githubusercontent.com/AvantiShri/model_storage/b6e1d69/deeplift/genomics/keras2_conv1d_record_5_model_PQzyq_modelWeights.h5
! [[ ! -f test.txt.gz ]] && wget https://raw.githubusercontent.com/AvantiShri/model_storage/9aadb769735c60eb90f7d3d896632ac749a1bdd2/deeplift/genomics/test.txt.gz
--2018-11-10 21:05:17-- https://raw.githubusercontent.com/AvantiShri/model_storage/db919b12f750e5844402153233249bb3d24e9e9a/deeplift/genomics/sequences.simdata.gz
Resolving raw.githubusercontent.com... 151.101.200.133
Connecting to raw.githubusercontent.com|151.101.200.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 629502 (615K) [application/octet-stream]
Saving to: 'sequences.simdata.gz'
100%[======================================>] 629,502 3.10MB/s in 0.2s
2018-11-10 21:05:18 (3.10 MB/s) - 'sequences.simdata.gz' saved [629502/629502]
--2018-11-10 21:05:18-- https://raw.githubusercontent.com/AvantiShri/model_storage/b6e1d69/deeplift/genomics/keras2_conv1d_record_5_model_PQzyq_modelJson.json
Resolving raw.githubusercontent.com... 151.101.200.133
Connecting to raw.githubusercontent.com|151.101.200.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 3010 (2.9K) [text/plain]
Saving to: 'keras2_conv1d_record_5_model_PQzyq_modelJson.json'
100%[======================================>] 3,010 --.-K/s in 0s
2018-11-10 21:05:18 (19.3 MB/s) - 'keras2_conv1d_record_5_model_PQzyq_modelJson.json' saved [3010/3010]
--2018-11-10 21:05:18-- https://raw.githubusercontent.com/AvantiShri/model_storage/b6e1d69/deeplift/genomics/keras2_conv1d_record_5_model_PQzyq_modelWeights.h5
Resolving raw.githubusercontent.com... 151.101.200.133
Connecting to raw.githubusercontent.com|151.101.200.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 153864 (150K) [application/octet-stream]
Saving to: 'keras2_conv1d_record_5_model_PQzyq_modelWeights.h5'
100%[======================================>] 153,864 --.-K/s in 0.05s
2018-11-10 21:05:19 (2.77 MB/s) - 'keras2_conv1d_record_5_model_PQzyq_modelWeights.h5' saved [153864/153864]
--2018-11-10 21:05:19-- https://raw.githubusercontent.com/AvantiShri/model_storage/9aadb769735c60eb90f7d3d896632ac749a1bdd2/deeplift/genomics/test.txt.gz
Resolving raw.githubusercontent.com... 151.101.200.133
Connecting to raw.githubusercontent.com|151.101.200.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 2287 (2.2K) [application/octet-stream]
Saving to: 'test.txt.gz'
100%[======================================>] 2,287 --.-K/s in 0s
2018-11-10 21:05:19 (17.6 MB/s) - 'test.txt.gz' saved [2287/2287]
Load the data
[3]:
! pip install simdna
Requirement already satisfied: simdna in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (0.4.2)
Requirement already satisfied: numpy>=1.9 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from simdna) (1.14.6)
Requirement already satisfied: scipy in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from simdna) (0.19.0)
Requirement already satisfied: matplotlib in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from simdna) (2.2.2)
Requirement already satisfied: subprocess32 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (3.5.2)
Requirement already satisfied: kiwisolver>=1.0.1 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (1.0.1)
Requirement already satisfied: six>=1.10 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (1.11.0)
Requirement already satisfied: python-dateutil>=2.1 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (2.7.3)
Requirement already satisfied: pytz in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (2018.5)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (2.2.0)
Requirement already satisfied: cycler>=0.10 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (0.10.0)
Requirement already satisfied: backports.functools-lru-cache in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from matplotlib->simdna) (1.5)
Requirement already satisfied: setuptools in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from kiwisolver>=1.0.1->matplotlib->simdna) (39.1.0)
You are using pip version 18.0, however version 18.1 is available.
You should consider upgrading via the 'pip install --upgrade pip' command.
[4]:
import simdna.synthetic as synthetic
import gzip
data_filename = "sequences.simdata.gz"
#read in the data in the testing set
test_ids_fh = gzip.open("test.txt.gz","rb")
ids_to_load = [x.decode("utf-8").rstrip("\n") for x in test_ids_fh]
data = synthetic.read_simdata_file(data_filename, ids_to_load=ids_to_load)
[5]:
import numpy as np
#this is set up for 1d convolutions where examples
#have dimensions (len, num_channels)
#the channel axis is the axis for one-hot encoding.
def one_hot_encode_along_channel_axis(sequence):
to_return = np.zeros((len(sequence),4), dtype=np.int8)
seq_to_one_hot_fill_in_array(zeros_array=to_return,
sequence=sequence, one_hot_axis=1)
return to_return
def seq_to_one_hot_fill_in_array(zeros_array, sequence, one_hot_axis):
assert one_hot_axis==0 or one_hot_axis==1
if (one_hot_axis==0):
assert zeros_array.shape[1] == len(sequence)
elif (one_hot_axis==1):
assert zeros_array.shape[0] == len(sequence)
#will mutate zeros_array
for (i,char) in enumerate(sequence):
if (char=="A" or char=="a"):
char_idx = 0
elif (char=="C" or char=="c"):
char_idx = 1
elif (char=="G" or char=="g"):
char_idx = 2
elif (char=="T" or char=="t"):
char_idx = 3
elif (char=="N" or char=="n"):
continue #leave that pos as all 0's
else:
raise RuntimeError("Unsupported character: "+str(char))
if (one_hot_axis==0):
zeros_array[char_idx,i] = 1
elif (one_hot_axis==1):
zeros_array[i,char_idx] = 1
onehot_data = np.array([one_hot_encode_along_channel_axis(seq) for seq in data.sequences])
Load the model
[6]:
from keras.models import model_from_json
#load the keras model
keras_model_weights = "keras2_conv1d_record_5_model_PQzyq_modelWeights.h5"
keras_model_json = "keras2_conv1d_record_5_model_PQzyq_modelJson.json"
keras_model = model_from_json(open(keras_model_json).read())
keras_model.load_weights(keras_model_weights)
Couldn't import dot_parser, loading of dot files will not be possible.
Using TensorFlow backend.
Install the deeplift package for the dinucleotide shuffling and visualzation code
[7]:
!pip install deeplift
Requirement already satisfied: deeplift in /Users/avantishrikumar/Research/clean_deeplift/deeplift (0.6.8.0)
Requirement already satisfied: numpy>=1.9 in /Users/avantishrikumar/anaconda/lib/python2.7/site-packages (from deeplift) (1.14.6)
You are using pip version 18.0, however version 18.1 is available.
You should consider upgrading via the 'pip install --upgrade pip' command.
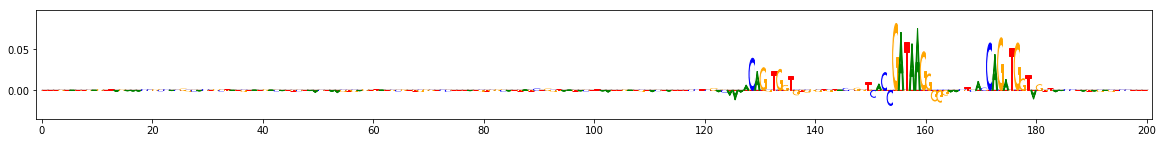
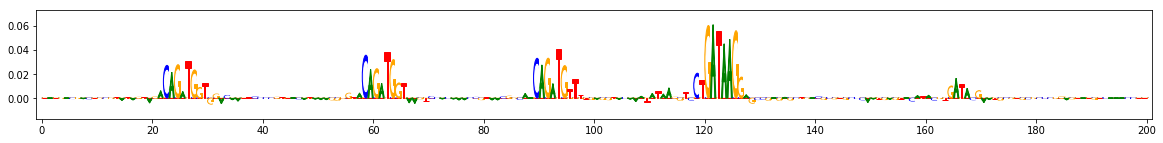
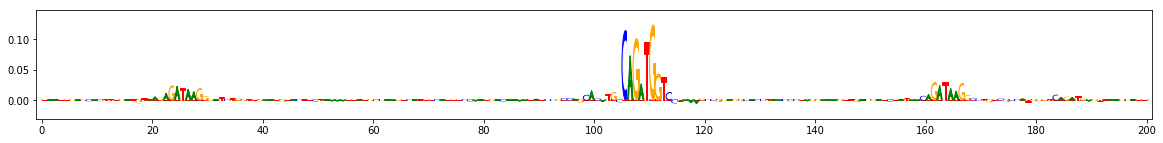
Compute importance scores¶
Define the function that generates the reference, in this case by performing a dinucleotide shuffle of the given input sequence
[8]:
from deeplift.dinuc_shuffle import dinuc_shuffle, traverse_edges, shuffle_edges, prepare_edges
from collections import Counter
def onehot_dinuc_shuffle(s):
s = np.squeeze(s)
argmax_vals = "".join([str(x) for x in np.argmax(s, axis=-1)])
shuffled_argmax_vals = [int(x) for x in traverse_edges(argmax_vals,
shuffle_edges(prepare_edges(argmax_vals)))]
to_return = np.zeros_like(s)
to_return[list(range(len(s))), shuffled_argmax_vals] = 1
return to_return
shuffle_several_times = lambda s: np.array([onehot_dinuc_shuffle(s) for i in range(100)])
Run DeepExplainer with the dynamic reference function
[9]:
from deeplift.visualization import viz_sequence
import shap
import shap.explainers.deep.deep_tf
reload(shap.explainers.deep.deep_tf)
reload(shap.explainers.deep)
reload(shap.explainers)
reload(shap)
import numpy as np
np.random.seed(1)
import random
seqs_to_explain = onehot_data[[0,3,9]] #these three are positive for task 0
dinuc_shuff_explainer = shap.DeepExplainer((keras_model.input, keras_model.output[:,0]), shuffle_several_times)
raw_shap_explanations = dinuc_shuff_explainer.shap_values(seqs_to_explain)
#project the importance at each position onto the base that's actually present
dinuc_shuff_explanations = np.sum(raw_shap_explanations,axis=-1)[:,:,None]*seqs_to_explain
for dinuc_shuff_explanation in dinuc_shuff_explanations:
viz_sequence.plot_weights(dinuc_shuff_explanation, subticks_frequency=20)
[ ]: