Partition explainer¶
This notebook demonstrates how to use the Partition explainer, which is an algorithm that uses a hierarchical clustering of the data to recursively partition the input space.
Tabular data example¶
By default the shap.Explainer interface uses the Parition explainer algorithm only for text and image data, for tabular data the default is to use the Exact or Permutation explainers (depending on how many input features are present). The reason the permutation explainer is preferred over the Partition explainer for tabular data is that the partition explainer algorithm will often leave credit on internal nodes of the hierarchical clustering tree. This works well for text and image data
where allocating credit to a contiguous chunk of the input makes sense. But for tabular data this credit sharing between input features in the same cluster can be undesirable. In contrast the Exact and Permutation explainers can still respect a hierarchical clustering tree of the input features while never leaving credit on internal nodes. Note that setting max_evals to infinity will make the Exact explainer, the Permutation explainer, and the Partition explainer all give the same answer, so
the tradeoffs here are all about how the SHAP values are approximated with a limited number of samples.
[1]:
import xgboost
import shap
# train XGBoost model
X,y = shap.datasets.adult()
model = xgboost.XGBClassifier().fit(X, y)
def f(x):
return model.predict(x, output_margin=True)
# compute SHAP values for the first 500 samples
bg = shap.maskers.Partition(shap.utils.sample(X,100))
explainer = shap.explainers.Partition(f, bg)
shap_values = explainer(X[:500])
explainers.Partition is still in an alpha state, so use with caution...
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-1-34a8e0adca24> in <module>
12 bg = shap.maskers.Partition(shap.utils.sample(X,100))
13 explainer = shap.explainers.Partition(f, bg)
---> 14 shap_values = explainer(X[:500])
~/projects/shap/shap/explainers/_explainer.py in __call__(self, max_evals, main_effects, error_bounds, batch_size, silent, *args, **kwargs)
188 row_result = self.explain_row(
189 *row_args, max_evals=max_evals, main_effects=main_effects, error_bounds=error_bounds,
--> 190 batch_size=batch_size, silent=silent, **kwargs
191 )
192 values.append(row_result.get("values", None))
~/projects/shap/shap/explainers/_partition.py in explain_row(self, max_evals, main_effects, error_bounds, batch_size, silent, *row_args)
452 output_indexes = None
453 fixed_context = 1
--> 454 output_indexes, base_value = self.owen(fm, max_evals // 2, output_indexes, fixed_context, batch_size, silent)
455
456 if False:
~/projects/shap/shap/explainers/_partition.py in owen(self, fm, npartitions, output_indexes, fixed_context, batch_size, silent)
497 M = len(fm)
498 m00 = np.zeros(M, dtype=np.bool)
--> 499 f00 = fm(m00.reshape(1,-1))[0]
500 base_value = f00
501 f11 = fm(~m00.reshape(1,-1))[0]
~/projects/shap/shap/utils/_masked_model.py in __call__(self, masks, batch_size)
51 return self._delta_masking_call(masks, batch_size=batch_size)
52 else:
---> 53 return self._full_masking_call(masks, batch_size=batch_size)
54
55 def _full_masking_call(self, masks, batch_size=None):
~/projects/shap/shap/utils/_masked_model.py in _full_masking_call(self, masks, batch_size)
119
120 joined_masked_inputs = self._stack_inputs(all_masked_inputs)
--> 121 outputs = self.model(*joined_masked_inputs)
122 _assert_output_input_match(joined_masked_inputs, outputs)
123
~/projects/shap/shap/explainers/_partition.py in <lambda>(x)
80 self.output_names = output_names
81
---> 82 self.model = lambda x: np.array(model(x))
83 self.expected_value = None
84 if getattr(self.masker, "clustering", None) is None:
<ipython-input-1-34a8e0adca24> in f(x)
7
8 def f(x):
----> 9 return model.predict(x, output_margin=True)
10
11 # compute SHAP values for the first 500 samples
~/anaconda3/lib/python3.7/site-packages/xgboost/sklearn.py in predict(self, data, output_margin, ntree_limit, validate_features, base_margin)
885 output_margin=output_margin,
886 ntree_limit=ntree_limit,
--> 887 validate_features=validate_features)
888 if output_margin:
889 # If output_margin is active, simply return the scores
~/anaconda3/lib/python3.7/site-packages/xgboost/core.py in predict(self, data, output_margin, ntree_limit, pred_leaf, pred_contribs, approx_contribs, pred_interactions, validate_features, training)
1440
1441 if validate_features:
-> 1442 self._validate_features(data)
1443
1444 length = c_bst_ulong()
~/anaconda3/lib/python3.7/site-packages/xgboost/core.py in _validate_features(self, data)
1852
1853 raise ValueError(msg.format(self.feature_names,
-> 1854 data.feature_names))
1855
1856 def get_split_value_histogram(self, feature, fmap='', bins=None,
ValueError: feature_names mismatch: ['Age', 'Workclass', 'Education-Num', 'Marital Status', 'Occupation', 'Relationship', 'Race', 'Sex', 'Capital Gain', 'Capital Loss', 'Hours per week', 'Country'] ['f0', 'f1', 'f2', 'f3', 'f4', 'f5', 'f6', 'f7', 'f8', 'f9', 'f10', 'f11']
expected Occupation, Education-Num, Capital Gain, Sex, Country, Marital Status, Race, Capital Loss, Age, Workclass, Hours per week, Relationship in input data
training data did not have the following fields: f3, f10, f0, f9, f7, f1, f6, f5, f4, f8, f2, f11
[3]:
import xgboost
import shap
# train XGBoost model
X,y = shap.datasets.adult()
model = xgboost.XGBClassifier().fit(X, y)
def f(x):
return model.predict(x, output_margin=True)
# compute SHAP values for the first 500 samples
bg = shap.maskers.Partition(shap.utils.sample(X,100))
explainer = shap.Explainer(f, bg)
shap_values = explainer(X[:500])
[7]:
shap.plots.bar(shap_values)
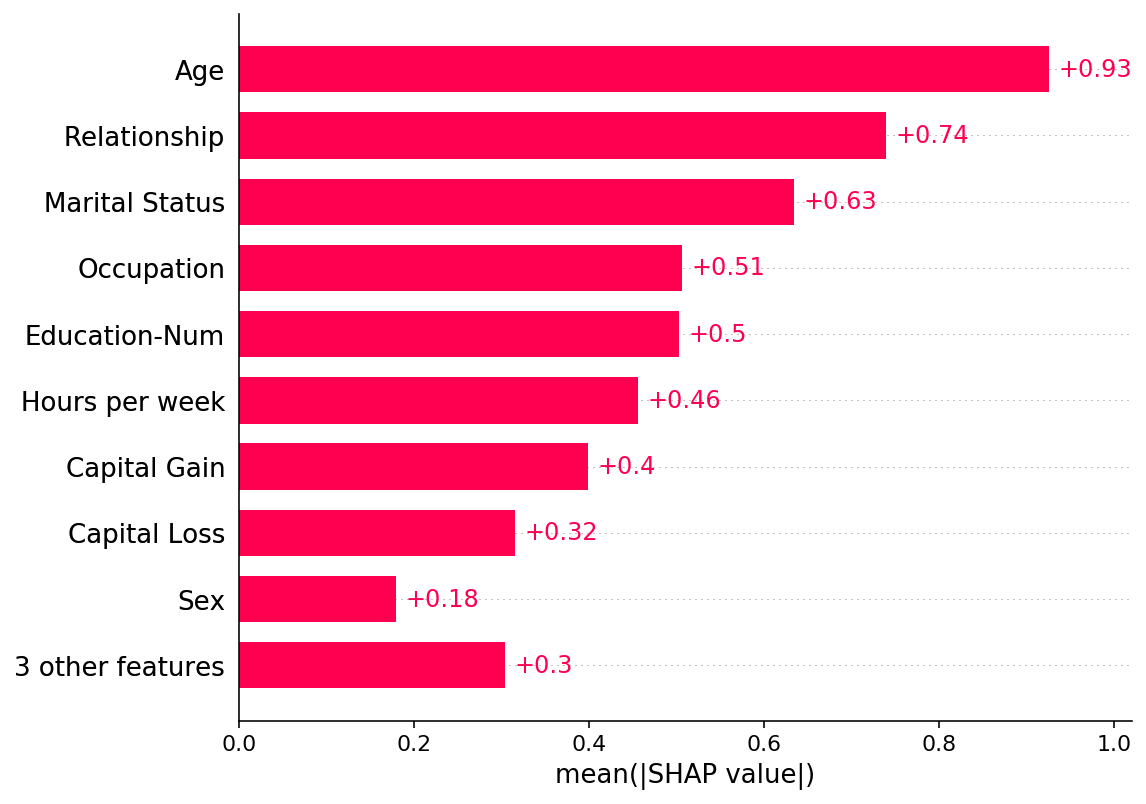
[6]:
shap.plots.bar(shap_values)
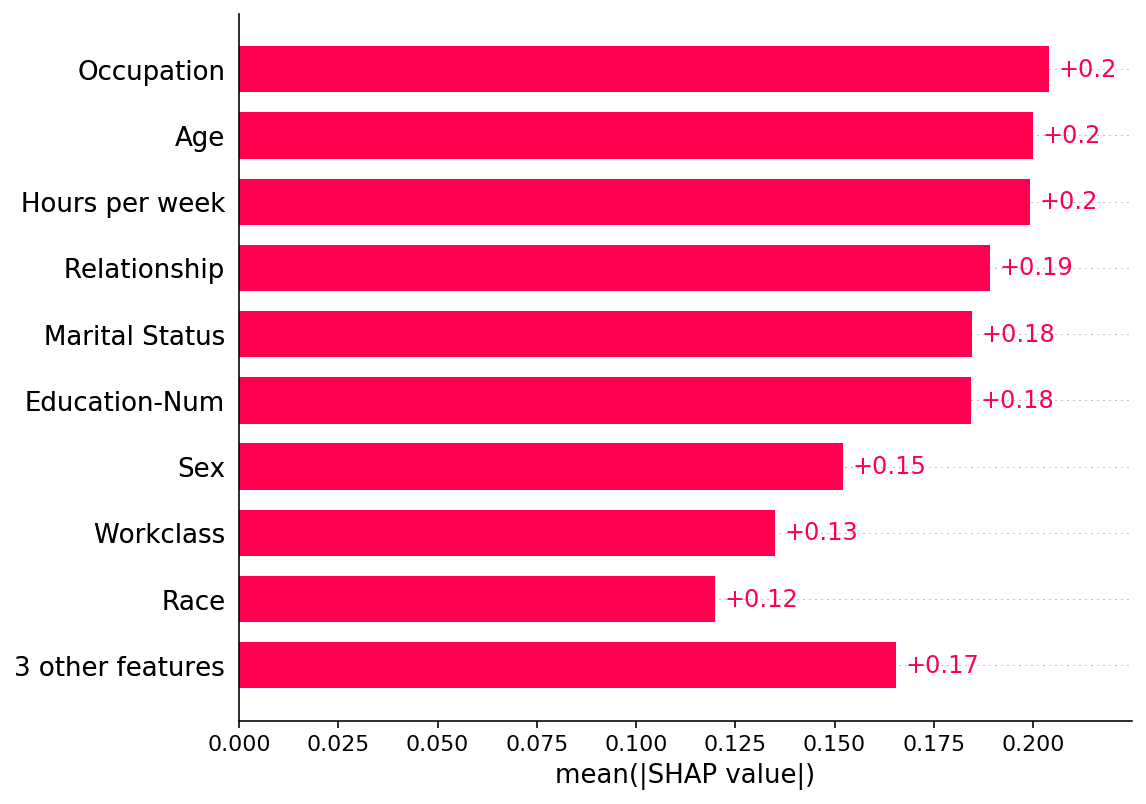
[5]:
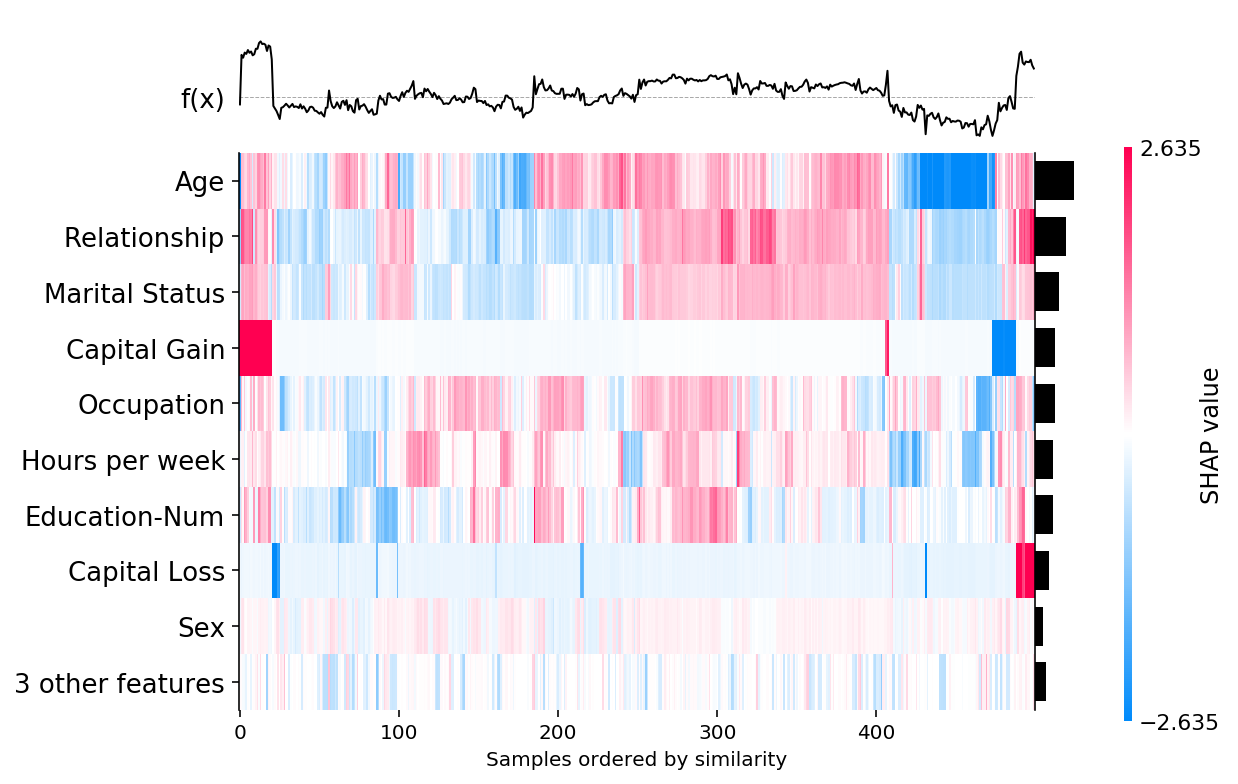
shap.plots.heatmap(shap_values)
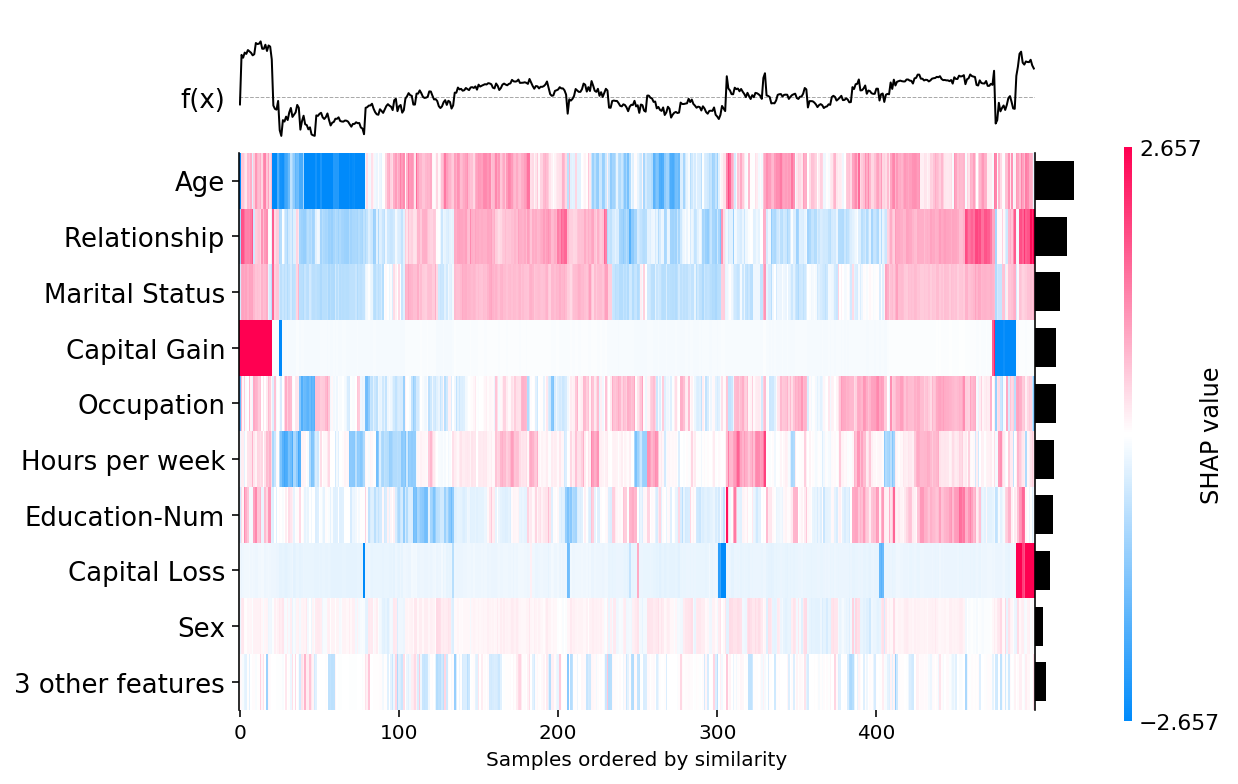
[13]:
shap.plots.heatmap(shap_values, instance_order=shap.order.sum)
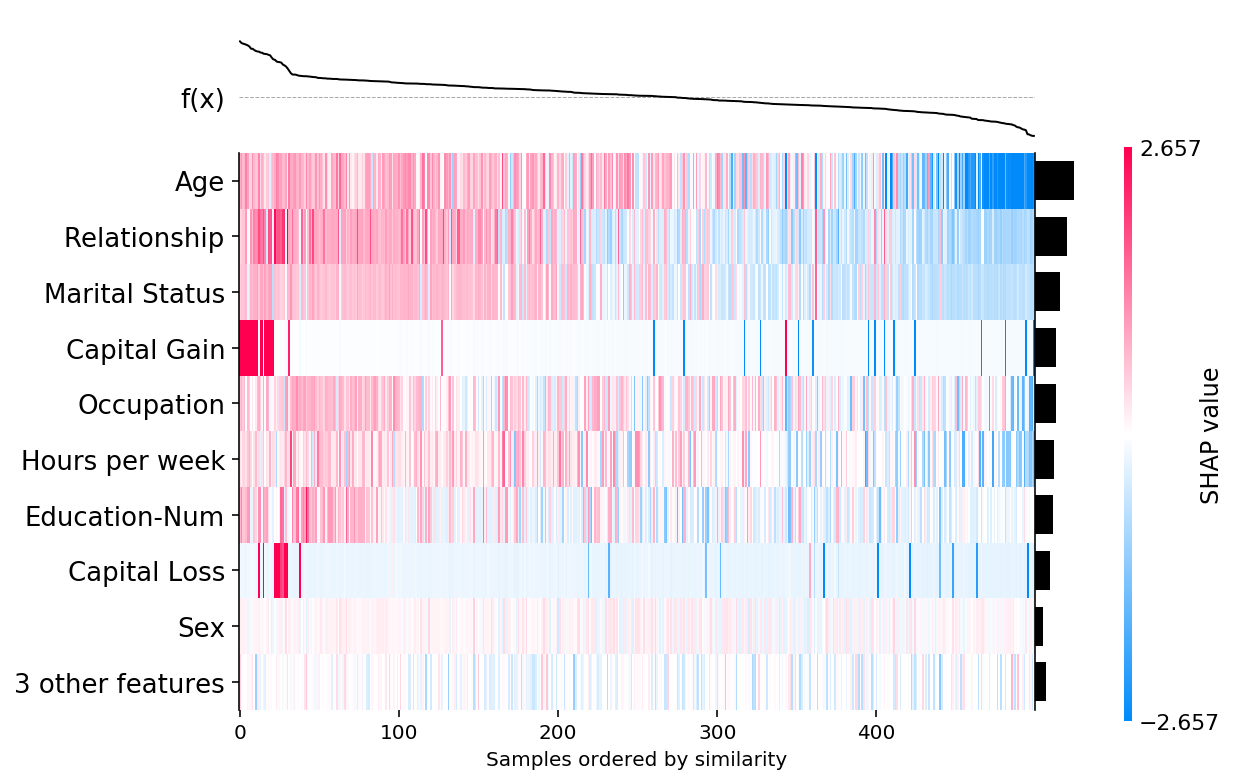
[9]:
shap.plots.heatmap(shap_values)
[ ]:
[ ]:
shap.maskers.Par
[ ]: